Inside Angle
From 3M Health Information Systems
AMIA 2017 Learning Showcase: Terminology-enabled clinical natural language processing for unstructured information extraction
The American Medical Informatics Association (AMIA) organizes a Learning Showcase during the Annual Symposium. Separate from the scientific program, the Learning Showcase allows exhibitors to present the informatics aspects or topics that relate to their products or services, avoiding a marketing or sales focus. In this context, I gave a talk on how terminologies can be used to help clinical natural language processing (NLP) during the latest AMIA Annual Symposium in November 2017, and would like to share the salient points here.
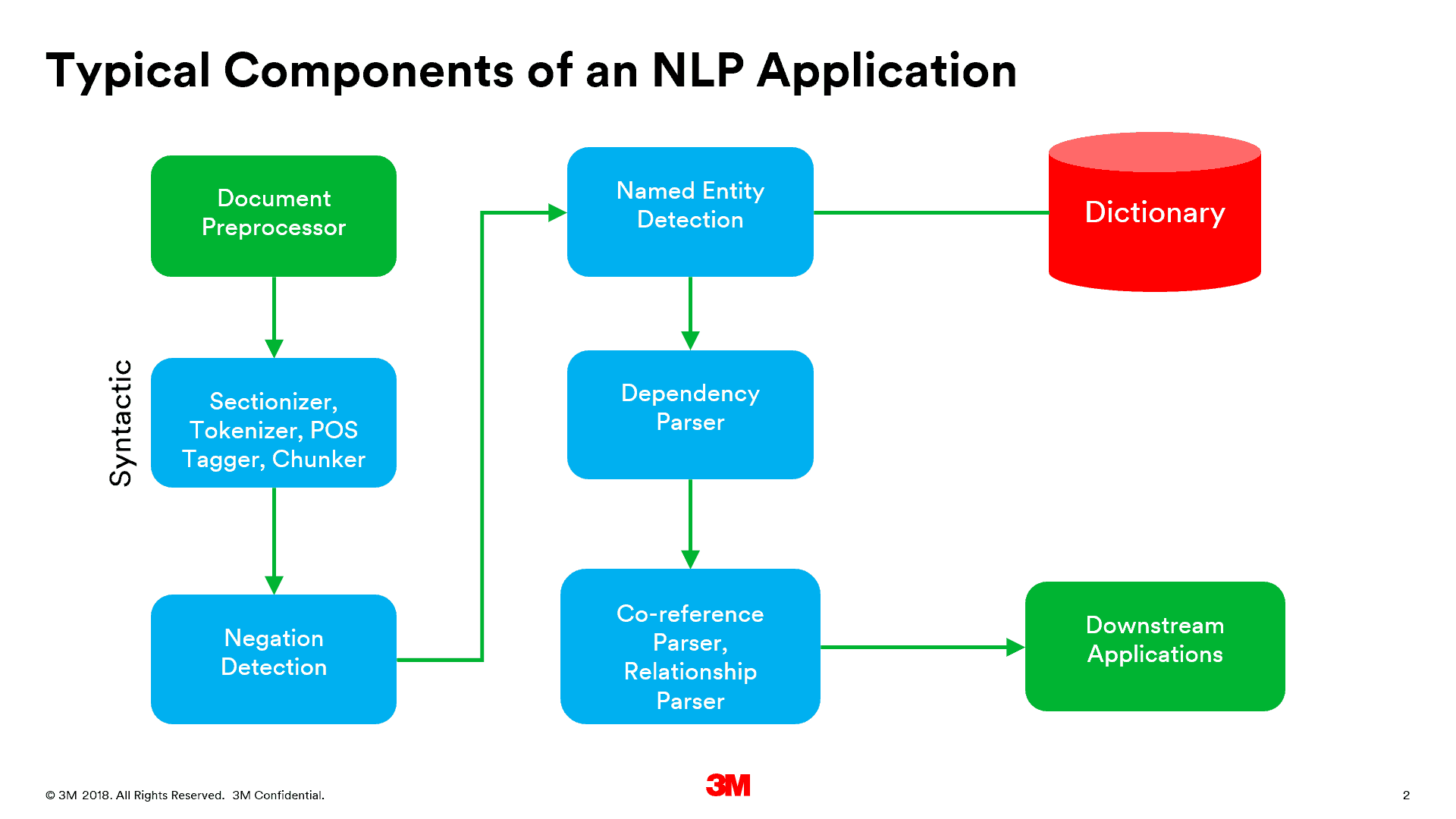
Typically, NLP pipelines include a syntactic parse followed by concept recognition and detection of semantic relationships. Syntactic parsing often consists of regioning, sentence detection, and parts of speech detection. Following this step, various named entity recognizers (NER) are used to identify concepts belonging to various semantic types and tag them with concept identifiers. Co-reference detection, negation detection and word sense disambiguation are done to resolve pronouns or atoms of a molecular concept, differentiate between assertations and negations, and disambiguate homonyms. These are done before or after NER as separate steps or by the NER component itself. An example of a typical NLP component pipeline is shown in the figure below:
Regardless of variations in the sequence of these annotators in the NLP pipeline, named entity recognition depends on domain-specific terminologies and terminology/ontology models to recognize concepts that may not be represented in a contiguous span of text. The variations in the pipeline and the NERs are often a result of whether the NLP implementation is application-specific or general purpose. Many initial applications of NLP in medicine have been application specific, designed to detect narrow types of information, e.g. smoking status, cardiac ejection fraction, etc. However, many general purpose clinical NLP applications are beginning to emerge, aimed at general purpose data mining, clinical documentation improvement, or information extraction from written documents, sometimes in combination with speech recognition of dictated clinical notes. Both offline and online NLP applications are beginning to emerge, with the online applications often combined with speech recognition to capture concepts and their relationships in clinical notes.
General purpose NLP applications have annotations designed to recognize a wide variety of concepts from various semantic types and use a broader vocabulary search space. Naturally, the initial trials of the generalizable applications are met with lower precision and recall compared to the application-specific NLP implementations. The breadth and depth of terminology content needed for general purpose clinical NLP can be a challenge. In addition, even when using terminologies such as SNOMED CT, which have a wide variety of concepts needed for clinical applications, implementers often need to subset, curate or enrich them to improve NLP performance. Traditionally, many NLP engines use a flat list of concept codes and descriptions exported from standard or local terminologies into their their NERs, which lead to reduced recall when the text spans containing the matching concepts are discontiguous, or there are differences due to pre- versus post-coordination between the terminology and the text. The rich terminology model of SNOMED CT can help to improve recall due to the pre- versus post-coordination issue, however, this comes at the cost of performance when millions of notes need to be processed. Furthermore, large reference terminologies such as SNOMED CT contain many homonyms that lead to reduced precision.
As a result, terminology management applications, such as the 3M Healthcare Data Dictionary (HDD), have been explored as a solution to provide the customized curation, subsetting and enrichment needed to use large standard terminologies needed for automated applications. For example, one requirement is to support creation of customized value sets needed for various NERs, such as the Heart Failure Value Set that contains concepts from SNOMED CT, ICD-10-CM and application-specific local terminologies. Specific NLP pipelines such as those for Registries can then be configured to use these value sets. In the absence of an intergrated terminology server, managing numerous value sets that contain overlapping concepts from a variety of standard and local terminologies is a laborious and error-prone process. In contrast, an integrated terminology server can hasten both the creation and maintenance of these value sets, and keep up with updated versions of the underlying terminologies. In addition, multilingual capabilities can be added independent of any terminologies, to provide a resource that’s available to be tapped when the NLP application is required to support other human languages.
NLP and standard terminologies have been long standing topics of research in medical informatics. Now, the focus should be on how we can apply the knowledge gained in working with terminologies to practical applications for NLP, specifically towards their performance and generalizability.
Senthil K. Nachimuthu, MD, PhD, medical informaticist with 3M Health Information Systems’ Healthcare Data Dictionary (HDD) team.